what does trypsin digest
 Trypsin digestion - YouTube
Trypsin digestion - YouTubeOur website does not fully support your browser. We have detected that you are using an earlier version of Internet Explorer. Your business experience can be limited. Please update your browser to Internet Explorer 11 or above. We believe that this site could serve you better: Singapore Language: EnglishPromega's Cookie Policy Our website uses functional cookies that do not collect any personal information or track your browsing activity. When selecting your country, you agree that we can place these functional cookies on your device. Your Account To protect your privacy, your account will be blocked after 6 failed attempts. After that, you will need to contact the Customer Service to unlock your account. You have four remaining attempts. You have three remaining attempts. You have two remaining attempts. You have one remaining attempt. To protect your privacy, your account has been blocked after 6 failed login attempts. Please contact the Customer Service to unlock your account. Username not found. This field is necessary. A password reset email has been sent to the main email address associated with your account. There was a problem with the process of restoring passwords. Please try again or contact the Customer Service. You have successfully restored your password. Your password reset link has expired. Please request another reset link. There was a problem that restored your password. Please try again or contact the Customer Service. You haven't checked your email address. A verified email address is required to access the full functionality of your Promega.com account. There was a problem that was recorded in his account. Please try again or contact the Customer Service. A verification email has been sent to the main email address associated with your account. There was a problem sending the verification email. Please try again or contact the Customer Service. Congratulations! You created a Promega.com account. Check your inbox to complete the email check. Note: You may not access your account until your email is verified. Our records indicate that this email address is already registered. There was a problem creating your account. Please try again or contact the Customer Service. There was a mistake processing your request. Please check your network settings and try again. Thank you for checking your email address. There was a problem checking your email address. Please try again or contact the Customer Service. Clinical Research " Diagnostics Drug Discovery Outstanding research topics Stocking insite Format and QC Automation solutions Custom Assay Development Articles, " Posters " " Tools " calculators Resources for students Events " Webinars " Literature reviewed by the Peer Technology Product use information Software & Firmware Global Support Our customer service and technical experts are here to help! Medical Affairs We provide medical information and facilitate research collaborations. Support local sales Contact a nearby dealer or sales representative. Your Cart Current Articles 0Protease Digestion for Masive Spectrometry This guide contains protocols for the digestion of proteins with trippsin, trippsin/lysC or alternative proteas. To request information about the products discussed here, visit our Proteas product pages. Mass Spectroscopy Analysis Mass spectrometry is a leading analytical method in proteomics (Mann et al., 2001). Mass spectrometry is used for the identification of proteins, the study of protein interactions:proteins, characterization of post-translational modifications (e.g. phosphorylation, glycosylation, methylation and acetylation) and quantification of proteins (relative and absolute). These applications use mainly proteomics from below, where the proteins of interest are digested with an enzyme such as trippsy and the resulting peptides are analyzed by mass spectrometry. The protein above focuses on the analysis of protein mixtures after the enzyme digestion of proteins in peptides. The complex mixture resulting from the peptides is analyzed using liquid chromatography in reverse phase (RP-LC) along with tandem mass spectrometry (MS/MS). The identification of peptides and later proteins are completed by combining fragmented ion spectra of peptides with theoretical spectra generated from protein databases. Trypsin has become the gold standard for protein digestion to peptides for shotgun proteomics. Trypsin's a proteasa serina. Hang proteins in peptides with an average size of 700-1500 daltons, which is in the ideal range for MS (Laskay et al., 2013). It is very specific, cutting on the carboxyl side of arginine and lysine residues. Arginine and C-terminal lysine peptides are loaded, making them detectable by MS. Trypsin is highly active and tolerant of many additives. Figure 1. Protheomic analysis from top to bottom with trippsin. Tripsy digestion protocolsThe specificity of trippsy activity is crucial for protein identification. However, specificity can be compromised by autolysis, which generates pseudotrypsin with an expanded specificity and an activity similar to chymotrypsin (Keil-Dlouha et al. 1971). Autolysis can result in additional peptide fragments that could interfere with database analysis and protein identification. Autolysis is suppressed by reduced methylation of lysine residues, producing a highly stable molecule (Rice et al. 1977). For maximum specificity, Promega offers Secuncing Grade Modified Trypsin (Cat. # V5111 and V5117), which is subject to reduction methylation and TPCK treatment to improve the specificity of cleavage of trippsin. TPCK inactivates the quimotrypsin activity. To further improve proteolytic efficiency, Promega developed a Grade of Tripsin Gold, Mass of Spectrometry (Cat. # V5280). More information and detailed protocol are available at Tripsin Gold, Mass Spectrometry Grade Technical Bulletin #TB309. In-Gel Protein Digestion There are harmful protocols for the digestion of in-gel proteins (Flannery et al. 1989; Shevchenko et al. 1996; Rosenfeld et al. 1992). Promega scientists have successfully used the following procedure. For a more simplified protocol, see the protocol for protein gel digestion using Trypsin and ProteaseMAXTM Surfactant, Tryspin Enhancer at ProteaseMAXTM Surfactant, Trypsin Enhancer, Technical Bulletin #TB373. Required materials:In-Solution Protein DigestionTypically, proteins are reduced and then rented to allow immediate access of the trippsy to the inner scot sites. This ensures high protein sequencing coverage during mass spectrometry analysis (Wilkinson, 1986). If high-sequence coverage is not required, the reduction and lease steps may be omitted. Note: To digest an unreduced protein, start this protocol in Step 3. Tripsin Gold, Mass Spec Grade Protocol More information and a detailed protocol for the use of Tripsin Gold, Mass Spec Grade are available in the Technical Bulletin #TB309. Affinity Tag In Vitro Pull-Down Assay with Trypsin Digestion and Protein AnalysisMarkillie and colleagues described a simple exogenous protein complex purification and identification method that can be easily automated (Markillie et al. 2005). The method uses MagneHisTM Ni particles (Cat.# V8560, V8565) to reduce target proteins, followed by denaturalizing elution, gut digestion and mass spectrometry analysis (Figure 2). Figure 2. Affinity label schematic diagram in a vitro drop-down test with trippsy digestion and mass spectrometry analysis. Trypsin/Lys-C Mix, MasaTrypsin/Lys-C Spectrum Grade (Cat.# V5071), is a preparation of guts with the highest protein efficiency. It's a mixture of Tripsin Gold, Mass Spectrometry Grade, and rLys-C, Mass Spec Grade. Proteolysis with Tripsin/Lys-C Mix, Grade Specter, generates proteptic peptides (i.e. peptides with C-terminal arginine and lysine residues). With the conventional tripsy digestion protocol (i.e. night incubation under non-denaturalizing conditions), Trypsin/Lys-C Mix improves protein digestion by eliminating most of the lost scoves (Figure 3). FIgure 3. Comparison side by side of the scote sites lost by trippsin or Tripsin/Lys-C Mix using a conventional digestion protocol The narrowly folded proteins represent a particular challenge; these proteins are resistant to proteolisis due to the inaccessibility of the internal scote sites for the trippsy. Using Trypsin/Lys-C Mix helps overcome this challenge by allowing a two-step digestion protocol (Figure 4), which uses the tolerance of Lys-C proteasa to the conditions of protein denaturalization. In step 1, a protein is denatured with 8M urea. In these conditions, Lys-C remains active and digests a protein in relatively large fragments. In the second step, the digestion mixture is diluted four times to reduce urea concentration to 2M. This reactivates the trippsin and allows complete proteolisis. Figure 4. Digestion of proteins difficult to digest using a two-step specialized procedure with Trypsin/Lys-C Mix.Trypsin/Lys C Protocol More information and protocol details are available on Tripsin/Lys-C Mix, Mass Spec Grade, Technical Manual #TM390. Alternative Protests The use of the tripasine in proteomics from below can impose certain limits on the ability to capture the complete proteoma. Double proteins can withstand the digestion of the trippsy. Post-translational modifications (PTMs) present a different challenge for trippsin because glucocans tend to limit access to scote sites, and acetylation causes lysine and arginine residues to resist trippsy digestion. To overcome these problems, the proteonomic community has begun to explore alternative proteas to complement the trippsin. However, protocols, as well as expected results generated in using these alternative proteas have not been systematically documented. Giansanti et. al. (2016) protocols optimized for six alternative proteas that have already shown promise in their applicability in proteomics, namely, chymotrypsin, Lys-C, Lys-N, Asp-N, Glu-C and Arg-C have been created. There are certain cases when trippsin does not provide adequate proteolisis. For example, many membrane proteins have a limited number of testeptic scote sites. In other cases, the distribution of scote sites is suboptimal, which results in peptides too long or too short for mass spectrometry analysis. Promega offers various alternative proteins that complement trippsin and allow efficient protein analysis with massive spectrometry. Figure 5 highlights the benefits of alternative protease quimotrypsin for protein mass spectrometry analysis. Figure 5. Increased protein coverage using both the trippsy and quimotrypsin. Site-Specific ProteasArg-C, Secuncing Grade (Cat.#), also known as clostripain, is an endopeptidase that accumulates in the C-terminus of arginine residue, including sites near proline. Arg... Activity C is optimal in pH 7.6-7.9. Asp-N, Sequencing Grade (Cat.# ) is an endoprotein that hydrolyzes peptide links on the N-terminal side of aspartic acid residues. The Asp-N activity is optimal in pH 4.0-9.0. rAsp-N, Mass Spec Grade (Cat.# ) is a recombinant protease that was cloned from Stenotrophomonas maltophilia and purified from E. coli. Chymotrypsin, Secuncing Grade (Cat.# ) is a seine endoprotein derived from the bovine pancreas. The proteasa hydrolyzes preferably on the carboxyl side of aromatic amino acids: thyrosine, phenylalanine and tryptophan. The activity of cytometry is optimal in pH 7.0-9.0.Glu-C, Secuncing Grade (Cat.# ) is a seine protein that is accumulated specifically on the C-terminal side of glutamic acid residues in the presence of ammonium bicarbonate or ammonia acetate. In phosphate bufers, the scote also occurs in aspartic acid residues. Glu- Activity C is optimal in pH 4.0-9.0.r Lys-C, Mass Spec Grade (Cat.# ) is endoproteinase recombinant Lys-C expressed in E. coli. The sequential origin of rLys-C is Protease IV of Pseudomonas aeruginosa. In the same way as the native Lys-C, rLys-C comes in the C-terminus of lysine residues with exceptional specificity and preserves proteolytic activity under denaturing conditions. rLys-C activity is optimal at pH 8.0-9.0. Endoproteinase Lys-C, Mass Spec Grade, (Cat.# ) is a highly purified protein seine that hydrolyzes specifically on the carboxyl side of the lysines. Lys... C maintains protein activity under strong conditions of protein denaturalization. Endoproteinase Lys-N, Mass Spec Grade, (Cat.# ) is a zinc metalloprotease that hangs on the amino side of the smoothies. Lys-N, Mass Spec Grado, preserves proteolytic activity under strong conditions of protein denaturalization such as 8M urea, Non-specific ProteasElastase (Cat.# ) is a seine protease that is preferred in the C-terminus of alanine, valine, serina, gluicillin, leucine or isoleucin residue. Elastase activity is optimal at pH 9.0. Pepsin (Cat.# ) Oriently clings to the C-terminus of phenylalanine, leucine, thyrosine and tritophan residues. Pepsine activity is optimal at pH 1.0-3.0. Thermolysin (Cat.# ) is a thermostable metalloproteins. Preferential thermolytes are agitated in the N-terminus of hydrophobic residues leucine, phenylalanine, valine, isoleucina, alanine and methionine. The optimal temperature range of digestion is 65 to 85° C. Thermolymine activity is optimal in pH 5.0-8.5. rLys-C LysC rLysN rAspN AspN ArgC GluC Cat. V1671 VA1170 VA1180 VA1160 V1621 V1881 V1651 Source and size Pseudomonas aeruginosa, expressed in E. coli (27.7kDa) Lysobacter enzymogenes (30kDa) Frondous tap (18kDa) Stenotrofomonas maltophilia (25kDa) Pseudomonas fragi (24.5kDa) Clostridium histolyticum (Subunits: 45kDa and 12kDa) Staphylococcus aureus V8 (27kDa) Cleavage sites Lys C-terminal. It doesn't clear if Lys is followed by Pro. Asp or Glu on the C-terminal side of Lys inhibits the neckline. Lys C-terminal. It doesn't clear if Lys is followed by Pro. Asp or Glu on the C-terminal side of Lys inhibits the neckline. Squares in Lysine's N-minus. Mainly on the N-terminal side of aspartic acid residues. The dung on the N-terminal side of glutamic acid residues can occur at a slower rate. N-terminal of Asp. C-terminal of Arg. It is also heard in Lys although at least efficiency. Glu C-terminal. Low-level scavages can occur in Asp residues also although at 100–300 doubles less efficiency. Protect:Protein Ratio (w/w) 1:20 to 1:50 1:20 to 1:100 1:20 to 1:100 1:10 to 1:100 1:20 to 1:200 1:20 to 1:350 1:20 to 1:200 Optimal pH range 8-9 hours pH 7–9 pH 7–9 rAsp-N has maximum activity at pH 8, but pH buffers ranging from 6-9 can be used. 4-9 pH 7.6-7.9 4-9 Reaction conditions 50–100mM Tris-HCl (pH 8) or 50mM NH4HCO3 (pH 7.8). Digestion at 37° C for 2-18 hours. Incubate 37° C for 2-18 hours. Incubate 37° C for 2-18 hours. Incubate 37° C for 60 minutes. 50mM Tris-HCl (pH 8). Digestion 2-18 hours at 37° C. 50mM Tris-HCl (pH 7.6-7.9), 5mM CaCl2, 2mM EDTA, √2mM DTT. Digestion 2-18 hours at 37° C. 100mM NH4HCO3 (pH 7.8), 50-100 mM HCL (pH 8). Digestion 2-18 hours at 37° C. Compatibility of buffer Tris-HCl, NH4HCO3 50mM Tris (pH 8) 50mM Tris (pH 8) Ammonium acetate (pH 5-6), HEPES (pH 7), Tris (pH 8-9), Ammonium bicarbonate (pH 8) Tris-HCl, NH4HCO3 Tris-HCl, NH4HCO3 NH4HCO3, ammonium acetate Compatibility of digestion in gel Yeah. Not tested Not tested Not tested Yeah. Yeah. Yeah. Compatibility ProteaseMAXTM Yeah. Not tested Not tested Not tested Yeah. Yeah. Yeah. Notes economic alternative to a native Lys-C proteasa. Similar to a native proteasa, rLys-C tolerates high denaturing conditions such as 8M urea. It is used to digest protein resistant proteins proteolyticly folded. It is also used as a trippsy alternative if the larger peptides are preferable for analysis. If urea is used in the preparation of the protein sample, avoid high temperatures. High temperature induces protein carbamylation in the presence of urea. It tolerates high denaturing conditions like 8M urea. It is used to digest protein resistant proteins proteolyticly folded. It is also used as a trippsy alternative if the larger peptides are preferable for analysis. If urea is used in the preparation of the protein sample, avoid high temperatures. High temperature induces protein carbamylation in the presence of urea. Lys-N is active in up to 6M urea, although the activity will vary depending on the concentration. If urea is used in the preparation of the protein sample, avoid high temperatures. High temperature induces protein carbamylation in the presence of urea. rAsp-N, Mass Spec Grade, is iyophilized in Tris (pH 8.0) with NaCl and stabilizing sugars. Rebuild in 50μl of ultra-pura water and mix gently. refurbished rAsp-N can be stored at 4°C for at least 8 weeks. For longer storage, one-use alibi can be stored at –65°C or below. Avoid cycles of defrosting. rAsp-N has a histidine label that can be used to remove it from the solution. It can be used as a trippsine alternative to achieve a better distribution of scotch sites. 100% of activity preserved in the presence of urea (up to 3.5 M), guanidine HCL (1M), SDS (up to 0.028%), ProteaseMaxTM Surfactant (up to 0.026%), acetonitrile (up to 60%), EDTA (up to 2mM); DTT or ß-mercaptoethanol. It is used in stone modification analysis. It requires DTT, cysteine or other reduction agent and CaCl2 for activity. It can be used as a trippsine alternative to achieve a better distribution of scotch sites. Glu- The activity C and the specificity of the neck is affected by the conditions of the buffer. In the ammonium bicarbonate and other non-phosphate buffers, Glu-C comes to Glu C-term. Glu- Cleaves at C-term Glu and Asp in phosphate buffer. More about alternative behaviorsArticle: Poster: You can learn more about using alternative proteas to improve coverage on-demand webinar: Bottoms Up: Improve the preparation of proteomic samples with recombinant Asp-N. ProteaseMaxTM Trypsin EnhancerProteaseMAXTM Surfactant, Trypsin Enhancer, allows fast and efficient digestion of in-solution and in-gel proteins such as trippsin and Lys-C. Surfing is an efficient protein-solubilization agent. (Figure 6). As an additive with urea, ProteaseMAXTM Surfactant improves the sun-ubilling effects of urea. Figure 6. Improved digestion of proteins in dissolution due to protein denaturalization using ProteaseMAXTM Surfactant ProteaseMAXTM Surfactant offers several advantages for in-gel protein digestion. First, this surfactant improves the identification of proteins through improved protein digestion and peptide extraction (Figure 7). Second, the surfactant minimizes the adsorption of peptides to plastic, which is the main cause of peptide loss during in-gel protein digestion. Third, ProteaseMAXTM Surfactant eliminates the need for post-digestion extraction and accelerates digestion. In the presence of ProteaseMAXTM In-gel protein digestion is complete in a single step of 1 hour. ProteaseMAXTM The surfactant is a surfactant anion that is degraded in the course of a digestion reaction, generating degradation products that are innocuous to mass spectrometry. This characteristic eliminates the need for post-digestion degradation. Figure 7. Example of better protein identifications when using ProteaseMAXTM Surfactant, Trypsin Enhancer, for elgel digestion of a complex protein sample. In-Gel Digestion of Proteins Using Trypsin and ProteaseMAXTM Surfactant, Trypsin Enhancer In-gel protein digestion saves time and work. The digestion passage is complete in 1 hour, and the ProteaseMAXTM Surfactant provides simultaneous extraction of gel peptides, eliminating the need for extraction of post-digestion peptides. The surfactant also improves the recovery of longer peptides that are normally preserved in the gel using a standard extraction protocol. For a detailed protocol, see the ProteaseMAXTM Surfactant, Trypsin Enhancer, Technical Bulletin #TB373. ProteaseMAXTM Protocol For more information on the use of ProteaseMAXTM Management of surgery and post-digestion peptides, see ProteaseMAXTM Surfactant, Trypsin Enhancer, Technical Bulletin #TB373. References Products and related resources for protein analysis CategoriesRelated GroupsGuides and Selectors Most important products Don't miss! Stay informed of Promega events, products and news. Do you need help? Local Contact Products " Resources " About Promega© 2021 Promega Corporation. All rights reserved. Asia Pacific AmericaEurope Don't you see your preferred location? The site will be displayed in English.

Rapid Trypsin Digestion of Complex Protein Mixtures for Proteomics Analysis | Sigma-Aldrich

Protease Digestion for Mass Spectrometry | Protein Digest Protocols
Trypsin - Worthington Enzyme Manual
AMINO ACID METABOLISM : DIGESTION / ABSORPTION

Digestion of Proteins: Grade 9 Understanding for IGCSE Biology 2.29 | PMG Biology

trypsin digestion Archives - Promega Connections

Trypsin - an overview | ScienceDirect Topics
Exocrine Secretions of the Pancreas

Trypsin: Definition, Function & Mechanism of Action | Study.com

Trypsin - Dodona
PRACTICAL 3 - THEORY
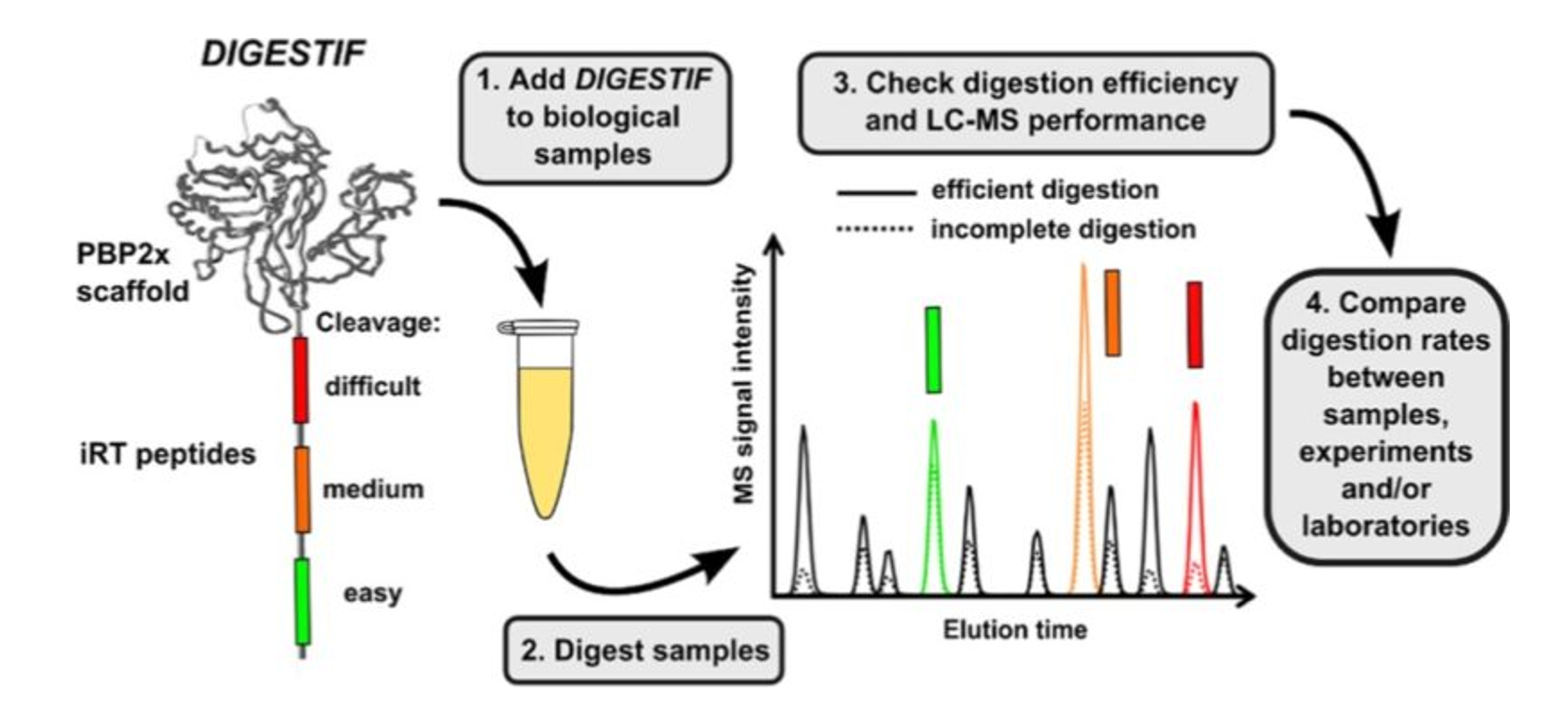
Quality Control of Tryptic Digestion

3.43 Protein Digestion in the Small Intestine | Nutrition Flexbook
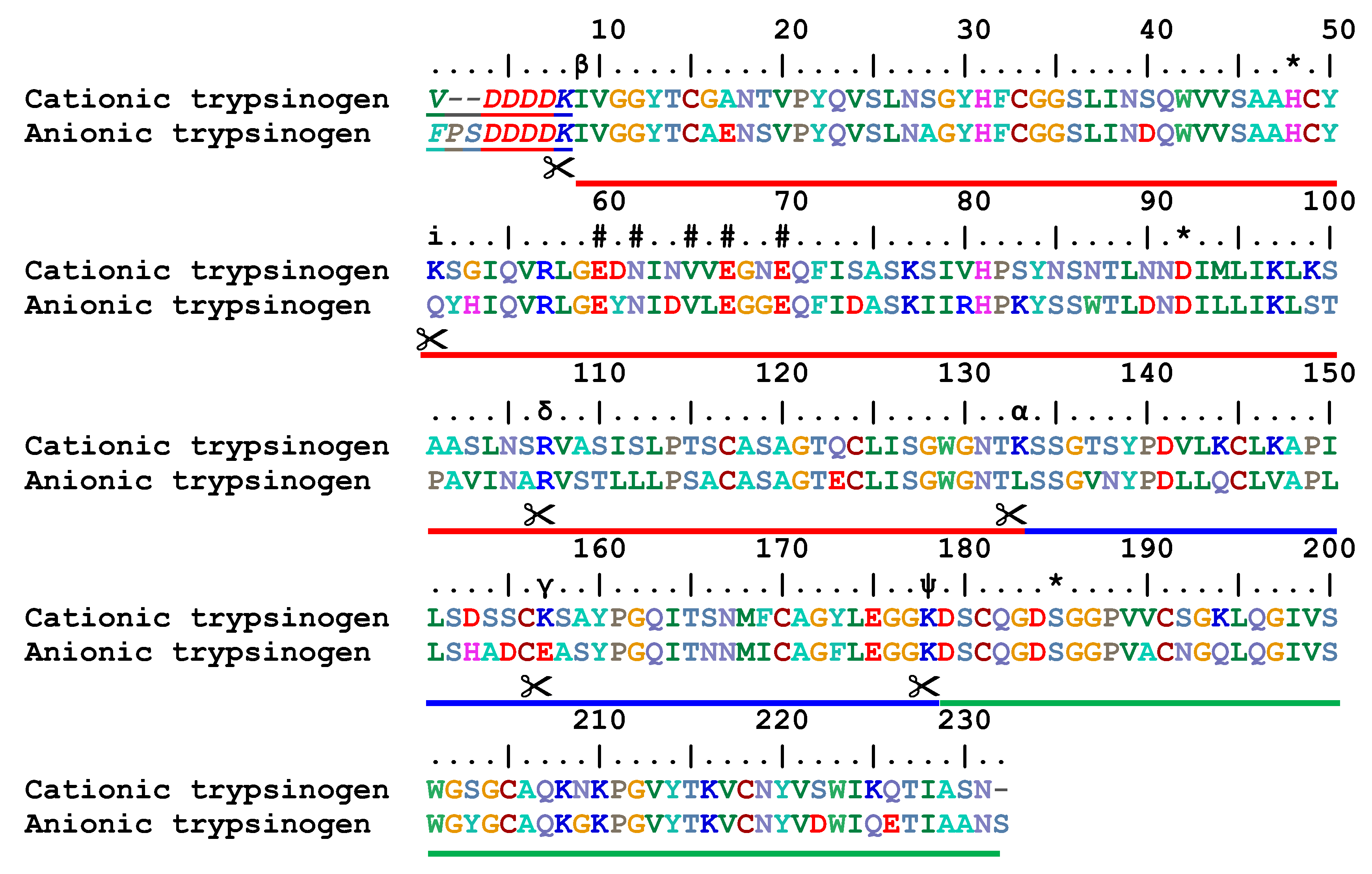
Molecules | Free Full-Text | Pseudotrypsin: A Little-Known Trypsin Proteoform | HTML
![PDF] Quantitative proteomics reveals the kinetics of trypsin-catalyzed protein digestion | Semantic Scholar PDF] Quantitative proteomics reveals the kinetics of trypsin-catalyzed protein digestion | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/cf2d639206d00563b33110d9d5f65ad7569421af/3-Figure1-1.png)
PDF] Quantitative proteomics reveals the kinetics of trypsin-catalyzed protein digestion | Semantic Scholar
If the proteases such as pepsin and trypsin, digest protein, why don't they digest the stomach and small intestine, since the stomach and small intestine are both made from protein? | Socratic
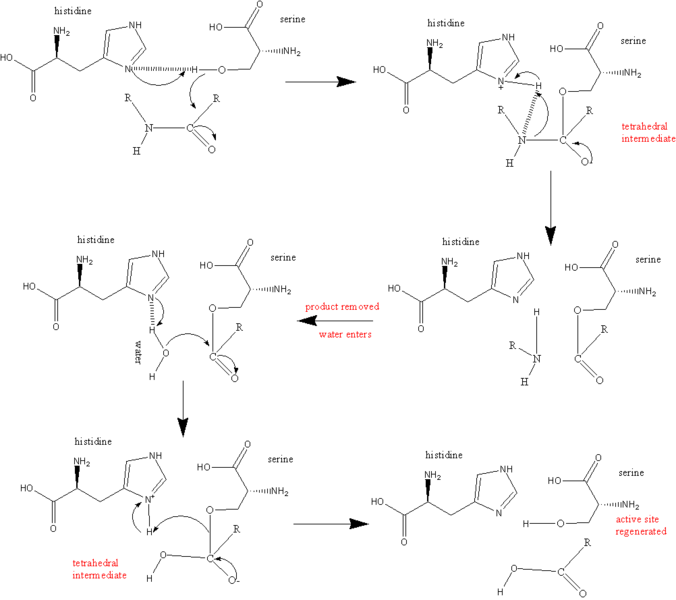
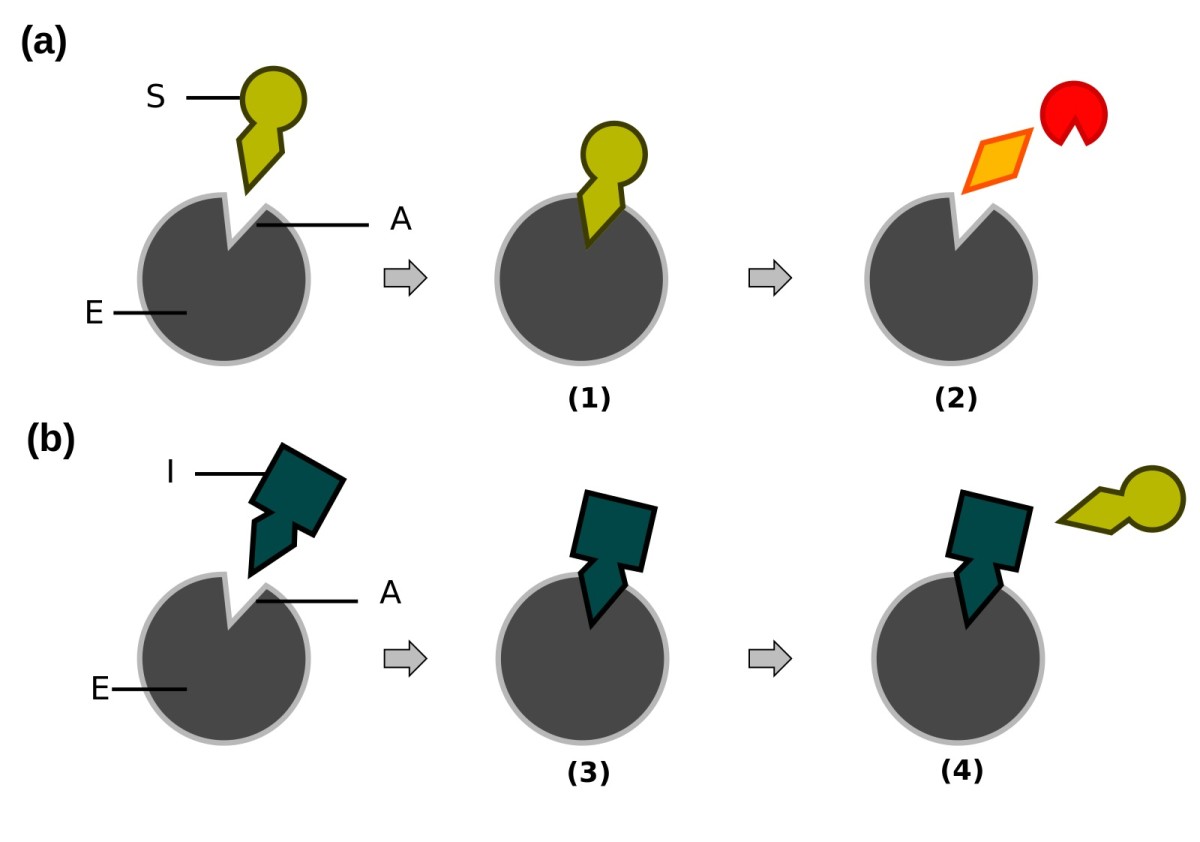
Enzyme Mechanisms: Serine Proteases

Development of a Simple and Rapid Digestion Protocol for Proteomics Sample Preparation | Sigma-Aldrich
Trypsin - Creative Enzymes
How Does Trypsin Work in Cell Culture - Pediaa.Com

Trypsin digestion | GoLectures | Online Lectures

Trypsin: Definition, Function & Mechanism of Action | Study.com
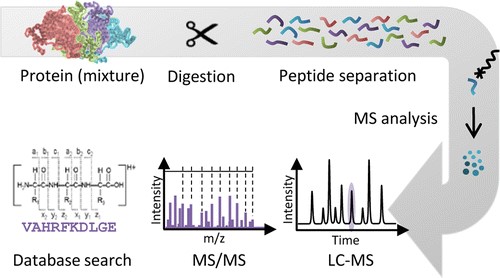
Protein Digestion Service - Creative Proteomics
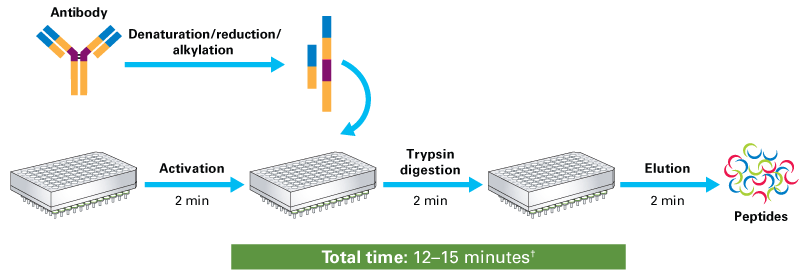
Protein and whole-cell lysate digestion with Capturem Trypsin technology

Same-Day Mass Spec Sample Prep Is Now a Reality: Shorten Digestion Time to as Little as 60 Minutes

Tryptic digestion of human serum for proteomic mass spectrometry automated by centrifugal microfluidics - Lab on a Chip (RSC Publishing) DOI:10.1039/D0LC00530D
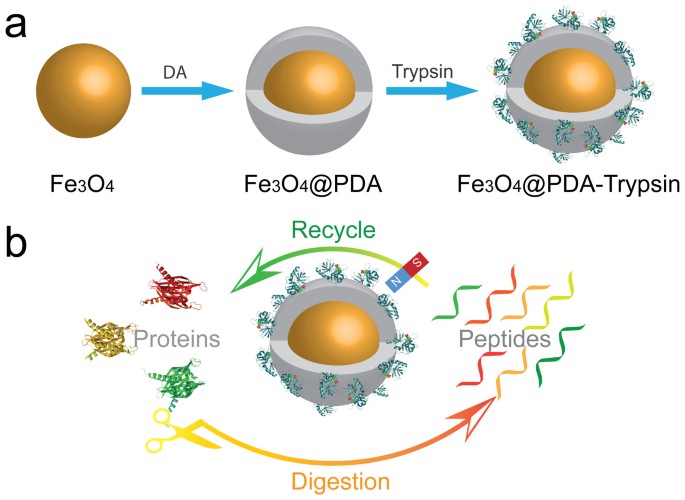
Construction of a high-performance magnetic enzyme nanosystem for rapid tryptic digestion | Scientific Reports
Mascot database search: Enzymes

Protease Digestion for Mass Spectrometry | Protein Digest Protocols

Protease Digestion for Mass Spectrometry | Protein Digest Protocols

Targeting proline in (phospho)proteomics - Laarse - 2020 - The FEBS Journal - Wiley Online Library

Sample Preparation Method for Accurate Analysis of Nonenzymatic PTMs in Biotherapeutic Proteins with Peptide Mapping

Manipulating trypsin digestion conditions to accelerate proteolysis and simplify digestion workflows in development of protein mass spectrometric assays for the clinical laboratory - ScienceDirect

Protease Digestion for Mass Spectrometry | Protein Digest Protocols
The Pancreas: Trypsin, Protein Digestion, and Pancreatitis - Owlcation - Education

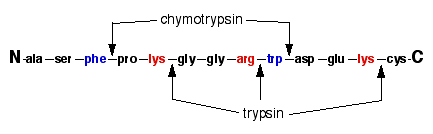
The Difference Between Trypsin & Chymotrypsin - Anatomy and Physiology Class | Study.com

Cell dissociation workflow with and without trypsin digestion. (A) The... | Download Scientific Diagram

Does anybody know if trypsin can cut in vitro at citrulline residues of a protein?
Automated Sample Preparation - 2015 - Wiley Analytical Science

Solved: Activity 2: Protein Digestion By Trypsin Trypsin H... | Chegg.com
Posting Komentar untuk "what does trypsin digest"